CPSF30 - a link between splicing, polyadenylation, and calcium/calmodulin signaling in plants
One of the subunits of the eukaryotic polyadenylation complex is a small protein, termed CPSF30 (for the mammalian protein) or Yth1p (the yeast counterpart). This protein consists of a characteristic array of conserved CCCH-type zinc finger proteins, binds RNA, and interacts with several other polyadenylation factor subunits. Arabidopsis possesses but one gene that might encode a CPSF30 homolog. Amino acid sequence comparisons suggest that the encoded protein is an authentic homolog of CPSF30 (5). Moreover, the Arabidopsis protein interacts with at least one other polyadenylation factor subunit, Fip1 (see the following; 4), and it can be immunoprecipitated from nuclear extracts with antibodies raised against the Arabidopsis CPSF100 protein (5). Interestingly, the Arabidopsis CPSF30 gene encodes two transcripts; the shorter of the two encodes a small polypeptide that resembles CPSF30 and Yth1p, while the longer of the two encodes a larger polypeptide that is essentially a fusion between CPSF30 and a domain that bears similarity to a poorly-characterized mammalian splicing factor (YT521-B; see Figure 1). This genomic organization of CPSF30 is conserved in rice, is also seen in Plasmodium, but does not occur in yeast or animals.
The Arabidopsis CPSF30 contains only three of the five zinc fingers that are found in other eukaryotic CPSF30 proteins, but is able to bind RNA much as does its eukaryotic counterparts. Remarkably, the Arabidopsis CPSF30 also binds calmodulin, and the RNA-binding activity of AtCPSF30 is inhibited by calmodulin in a calcium-dependent fashion (5). This observation suggests that mRNA 3' end formation in plants may be regulated directly by calmodulin, and thus by stimuli whose action is mediated by calcium/calmodulin signaling.
Together with our collaborators (Drs. Li and Falcone), we are characterizing a mutant with a T-DNA insertion in the first exon of At1g30460. While this mutant produces no detectable CPSF30 protein or mRNA (5), it is viable (but somewhat dwarfed in stature). The fact that CPSF30-deficient plants are viable suggests that factors other than or in addition to CPSF30 can replace this protein in polyadenylation. The identity of these alternative factors remains a mystery, and is one subject of current study.
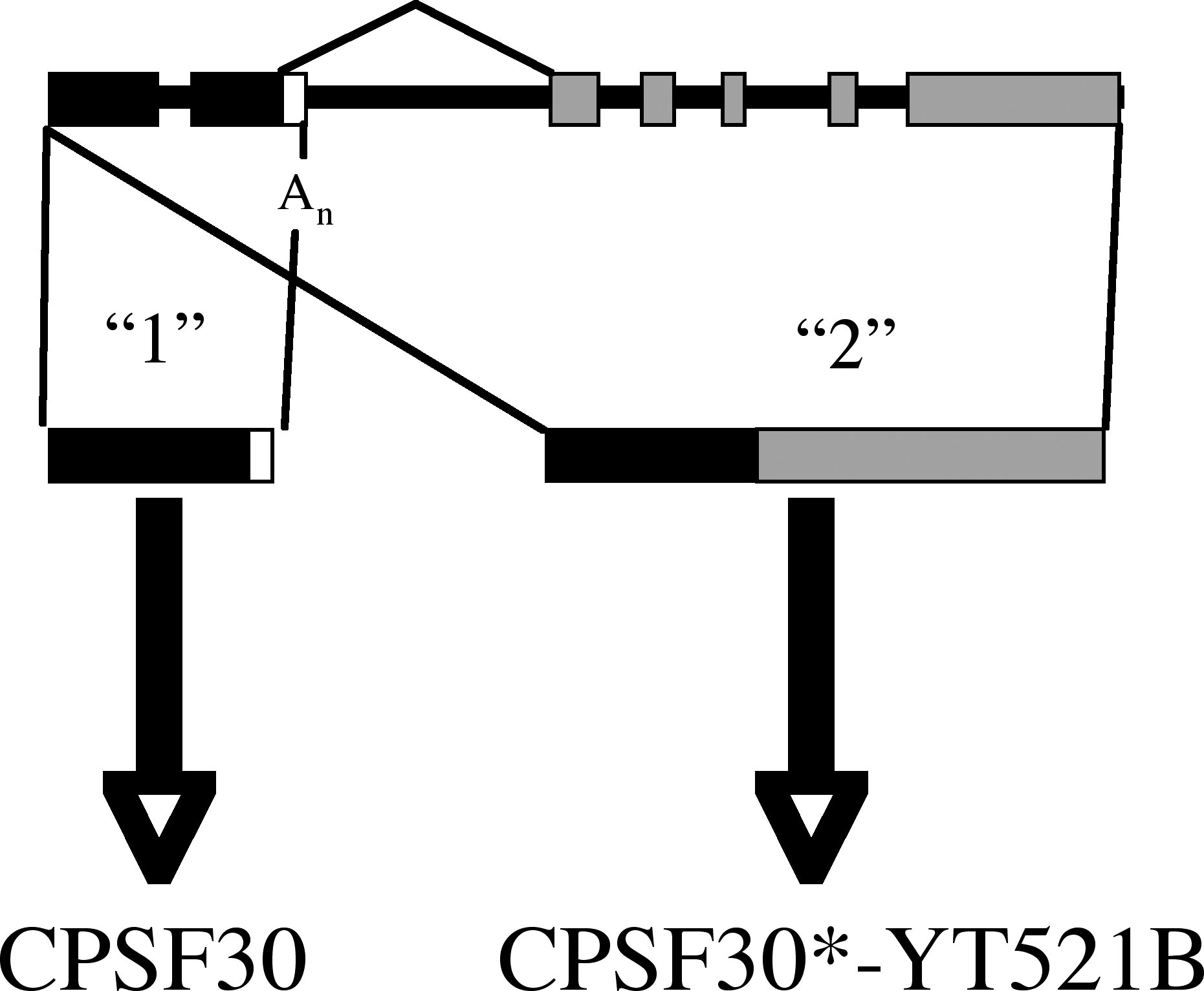
Figure 1. Illustration of the Arabidopsis gene (At1g30460) that encodes CPSF30. The structure of the genomic DNA is shown at the top, and the two transcripts that are derived by alternative RNA processing below. Transcript "1" encodes CPSF30, while transcript "2" encodes a novel CPSF30-YT521B fusion protein.
Fip1 - an interaction hub that links several polyadenylation factor subunits to poly(A) polymerase
One Arabidopsis gene whose product resembles a mammalian and yeast polyadenylation factor subunit is the so-called AtFip1(V) gene (4). The encoded protein bears a number of similarities to its human counteraprt - it binds RNA (with a modest preference for RNAs containing an FUE), it alters the activity of poly(A) polymerase, and it interacts with a number of other Arabidopsis polyadenylation factor subunits (see Figure 2). Among he latter are two (AtCFIm-25 and a nuclear poly(A) binding protein) that have not been reported to interact with Fip1 in other systems. These results suggest a model in which AtFip(V) is the center of a network of RNA-binding proteins and other factors that are involved in mRNA 3' end formation. They also provide coneptual links of a number of Arabidopsis proteins to poly(A) polymerase, thus strengthening the supposition that these proteins are authentic polyadenylation factor subunits.
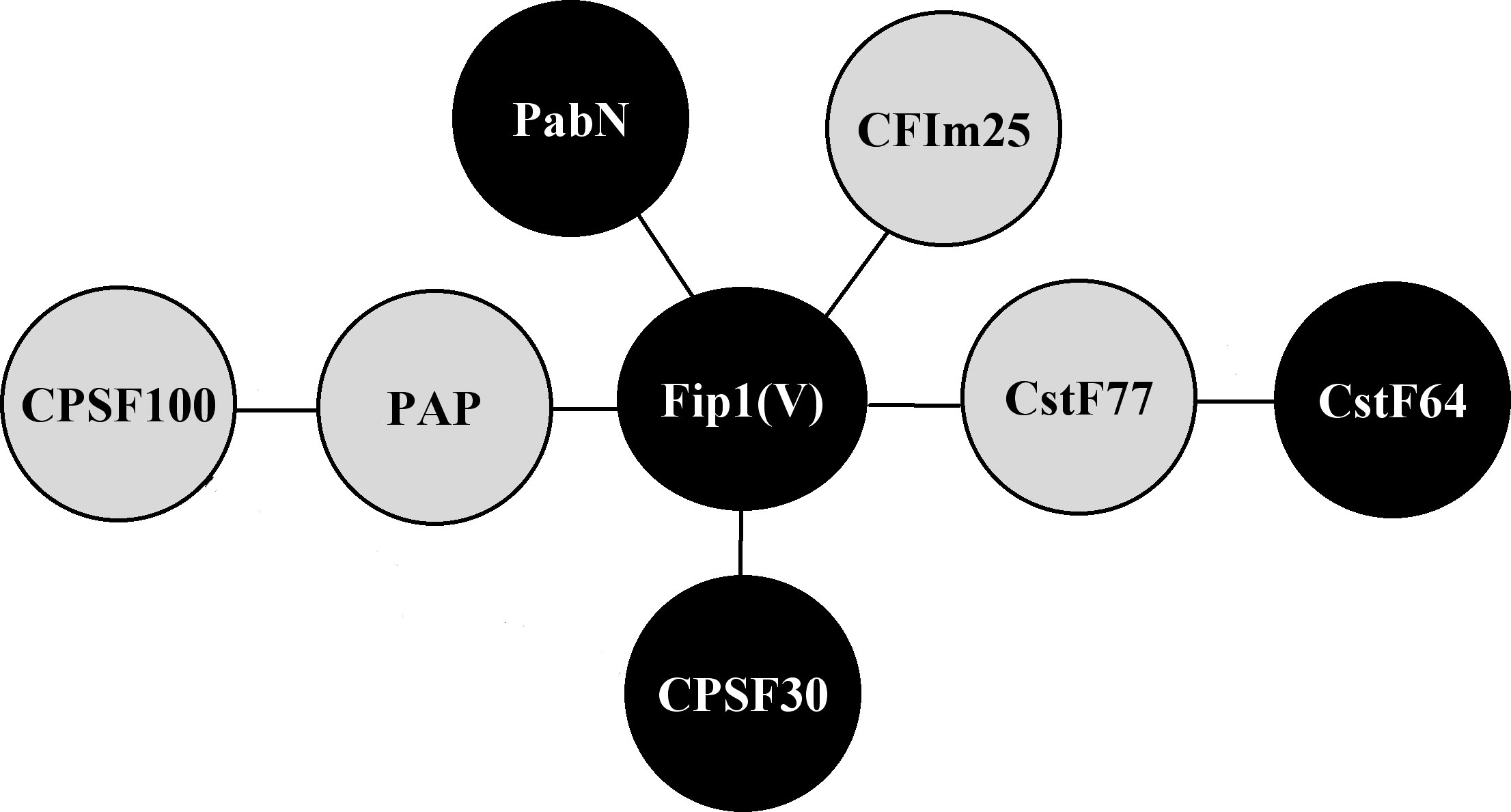
Figure 2. Summary of protein-protein interactions amongst Arabidopsis polyadenylation factor subunits that have been reported in the literature. Interactions are denoted as lines connecting the various subunits. The AtPAP-AtCPSF100 interaction was reported by Elliott et al. (2) and the AtCst77-AtCstF64 interaction by Yao et al. (6); the others are described in Forbes et al. (4). Black circles denote proteins that bind RNA.
Poly(A) polymerases - an interesting gene family
We have identified and partially purified a poly(A) polymerase from nuclei isolated from pea leaves (1), and we have identified Arabidopsis and rice genes whose predicted products are similar to mammalian polyadenylation factor subunits (including the poly(A) polymerase). Interestingly, the Arabidopsis CPSF100 homolog has been found to interact with at least one of the four Arabidopsis PAPs (2). Currently, this set of genes is being analyzed using a battery of approaches (see this site for details and progress).
The Arabidopsis genome has four genes that encode poly(A) polymerase isoforms (3). One of the four isoforms is much smaller than the other three, lacking the RNA-binding domain and nuclear localization signals that are hallmarks of canonical poly(A) polymerases (including the other three Arabidopsis isoforms). Remarkably, RNAs from all four genes are alternatively-spliced near the 5'-end of their respective transcription units, so that mRNAs encoding highly-truncated polypeptides are produced. This alternative splicing varies from tissue to tissue; along with tissue specificity in the expression of the four genes, this suggests that different poly(A) polymerase isoforms may play specific roles during plant development.
Bibliography:
1. Hunt AG, Meeks LR, Forbes KP, Das Gupta, Mogen BD. 2000. Nuclear and chloroplast poly(A) polymerases from plants share a novel biochemical property. Biochemical and Biophysical Research Communications 272, 174–181.
2. Elliott BJ, Dattaroy T, Meeks-Midkiff LR, Forbes KP, Hunt AG. 2003. An interaction between an Arabidopsis poly(A) polymerase and a homologue of the 100 kDa subunit of CPSF. Plant Mol. Biol. 51:373-84
3. Addepalli B, Meeks LR, Forbes KP, Hunt AG. 2004. Novel alternative splicing of mRNAs encoding poly(A) polymerases in Arabidopsis. Biochimica Biophysica Acta 1679: 117-128.
4. Forbes KP, Addepalli B, Hunt AG. 2006. An Arabidopsis Fip1 homologue interacts with RNA and provides conceptual links with a number of other polyadenylation factor subunits. J. Biol. Chem. 281: 176-186.
5. Delaney KJ, Xu R, Zhang J, Yun K-Y, Li QQ, Falcone DF, Hunt AG. 2006. Calmodulin interacts with and regulates the RNA-binding activity of an Arabidopsis polyadenylation factor subunit. Plant Physiol. 140: 1507-1521.
6. Yao Y, Song L, Katz Y, Galili G. 2002. Cloning and characterization of Arabidopsis homologues of the animal CstF complex that regulates 3' mRNA cleavage and polyadenylation. J. Exp. Bot. 53:2277-8.
Return to the Poly(A) Main Page
Return to the Art Hunt's Home Page