General characteristics of plant polyadenylation signals
This page contains three figures: an illustration of the general structure
of plant polyadenylation signals (Figure 1),
a representation of the base content between 1 and 60 nts upstream from
plant poly(A) sites (Figure 2), and a representation
of the base content between 5 nts upstream and 5 nts downstream from plant
poly(A) sites (Figure 3).
Figure 1
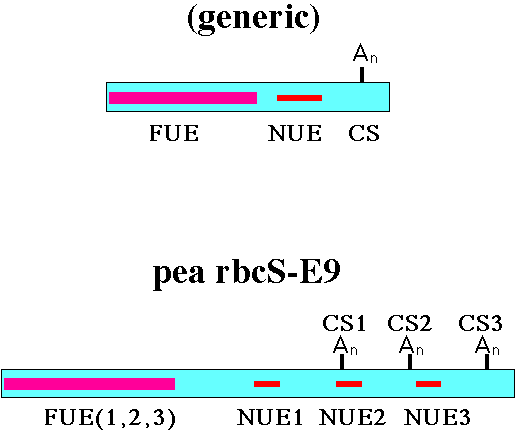
Figure 1. The general structures of a generic plant polyadenylation signal,
and of the pea rbcS-E9 gene (1,2), are shown here. FUE - Far-Upstream Element.
NUE - Near-Upstream Element. CS - polyadenylation/Cleavage Site. Note that,
as shown for the rbcS-E9 poly(A) signal, each CS is controlled by a separate,
distinct NUE (1,3), and all three sites are controlled by a single FUE (1).
Figure 2
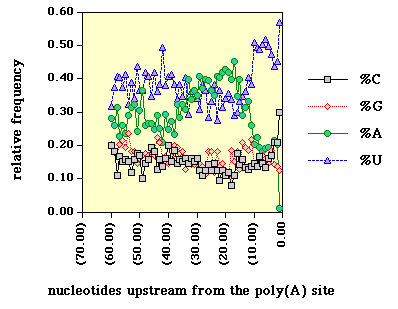
Figure 2. The sequences between 1 and 60 nt upstream from reported polyadenylation
sites in 211 plant genes were compiled and analyzed. Only those published
sequences where the 3' end(s) of the corresponding RNAs were established
by transcript mapping or by location of an extended polyadenylate tract
in a cDNA were included. In those instances where multiple polyadenylation
sites were reported, only the most distal site was included; although this
might result in the inadvertant inclusion of overlapping or near-overlapping
signals, there is no consistent spacing or periodicity of 3' termini in
those instances where multiple poly(A) sites have been reported (unpublished
observations). In all cases, nucleotide -1 was defined as the base immediately
preceding the first A in the polyadenylate tract. This may not be completely
accurate since most polyadenylate tracts in cDNAs occur at positions where
one or more adenines exist in the corresponding genomic clone; however,
since it is impossible to define which adenines are added during and which
after transcription, I have arbitrarily chosen the last non-A base as -1
in these cases. Although not exhaustive, the list of genes includes those
reported as early as 1982 and as late as 1990. The list of genes, the compiled
sequences, and references are available upon request.
Things to note here include the generally low G and C content, the
elevated A content between 10 and 40 nts upstream from the poly(A) site
(this probably reflects the high A content of NUEs [3]), and the pronounced
U and C content between 1 and 10 nts upstream from the poly(A) site.
Figure 3
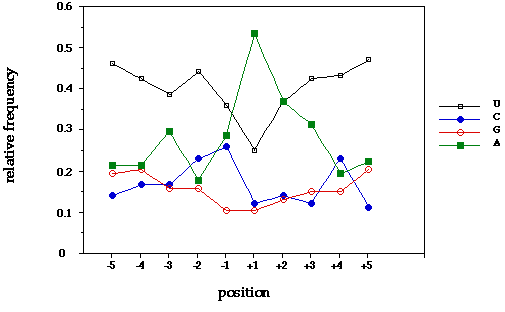
Figure 3. The 3' end cleavage sites analyzed here were derived from genomic
clones, or from cDNAs with reported 3' end heterogeneity, such that 3'-flanking
sequences for some sites were known. Cleavage sites were analyzed exactly
as reported. The notation -1 designates the base immediately preceding the
poly(A) tail. Likewise, +1 denotes the base immediately downstream from
"-1".
Things of significance here are the generally high U content, the
elevated C content at -2 and -1, and the abundance of A at or immediately
after the poly(A) site (-1).
References:
1. Mogen, B. D., MacDonald, M. H., Leggewie, G., and Hunt, A. G. (1992).
Several distinct types of sequence elements are required for efficient mRNA
3' end formation in a pea rbcS gene. Mol. Cell. Biol. 12, 5406-5414.
2. Hunt, A. G. (1994) Messenger RNA 3' end formation in plants. Ann. Rev.
Plant Physiol. Plant Mol. Biol. 45, 47-60.
3. Li, Q. and Hunt, A. G. (1995) A near upstream element in a plant polyadenylation
signal consists of more than six bases. Plant Mol. Biol. 28, 927-934.



