CPSF30 - a link between splicing, polyadenylation, and calcium/calmodulin signaling in plants
Overview - CPSF30 in plants and other eukaryotes
One of the subunits of the eukaryotic polyadenylation complex is a small protein, termed CPSF30 (for the mammalian protein) or Yth1p (the yeast counterpart). This protein consists of a characteristic array of conserved CCCH-type zinc finger proteins, binds RNA, and interacts with several other polyadenylation factor subunits. Arabidopsis possesses but one gene that might encode a CPSF30 homolog. Amino acid sequence comparisons suggest that the encoded protein is an authentic homolog of CPSF30 (1). Moreover, the Arabidopsis protein interacts with at least one other polyadenylation factor subunit, Fip1 (2), and it can be immunoprecipitated from nuclear extracts with antibodies raised against the Arabidopsis CPSF100 protein (1). The purified Arabidopsis protein possesses a distinctive endonuclease activity, suggestive of a role for this protein as a processing endonuclease in the polyadenylation reaction (3). In this respect, the Arabidopsis protein resembles the so-called Clipper protein from Drosophila (4,5). This observation raises interesting questions as to the nature of the 3' end processing reaction in plants; this follows from the reported activity of the Arabidopsis CPSF30 ortholog, the fact that the gene encoding this protein is both unique and non-essential in Arabidopsis (1), and reports that another polyadenylation factor subunit, CPSF73, may also be a 3' end-processing endonuclease (6,7).
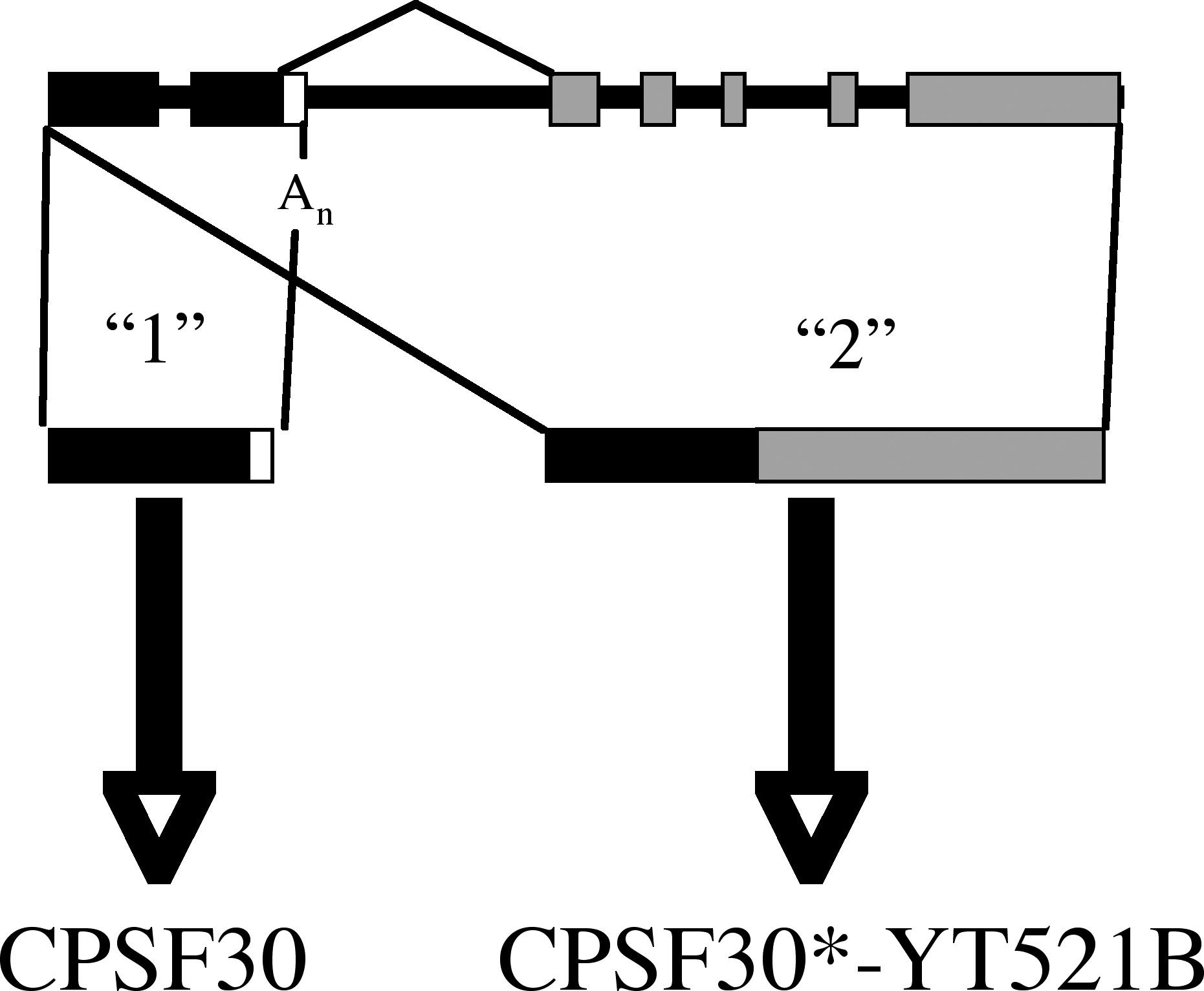
Interestingly, the Arabidopsis CPSF30 gene encodes two transcripts; the shorter of the two encodes a small polypeptide that resembles CPSF30 and Yth1p, while the longer of the two encodes a larger polypeptide that is essentially a fusion between CPSF30 and a domain that bears similarity to a family of proteins that are readers of a wide-ranging RNA modification (methylation at the N6 position of adenosines; the domain in the larger isoform of plant CPSF30 proteins is noted as YT521-B in Figure 1). This genomic organization of CPSF30 is conserved in many plants (15) but does not occur in yeast or animals.
Biochemical properties of the Arabidopsis CPSF30
The Arabidopsis CPSF30 contains only three of the five zinc fingers that are found in other eukaryotic CPSF30 proteins (Figure 2), but is able to bind RNA much as does its eukaryotic counterparts. Remarkably, the Arabidopsis CPSF30 also binds calmodulin, and the RNA-binding activity of AtCPSF30 is inhibited by calmodulin in a calcium-dependent fashion (1). This observation suggests that mRNA 3' end formation in plants may be regulated directly by calmodulin, and thus by stimuli whose action is mediated by calcium/calmodulin signaling.
As stated in the preceding, this protein possesses three zinc finger motifs. The first of these has been implicated in RNA binding, while the third is apparently the nuclease (3). Mutational analysis (3) and studies of the effects of salt and temperature on the protein (8) suggest that these two zinc finger motifs act independently, such that RNA binding is not needed for nuclease activity, and nuclease activity is not needed for RNA binding. The Arabidopsis CPSF30 requires zinc for RNA binding and nuclease activity, as the enzyme is inhibited by zinc chelators (8). However, relatively high concentrations of zinc, as well as cadmium, inhibit the nuclease activity (but not RNA binding; 8). The novel plant-specific N-terminus is needed for this inhibition; the similarity in amino acid composition between the N-termini of CPSF30 and AtFip1(V), which also inhibits CPSF30 nuclease activity, suggests that Zn in some way promotes or enables an interaction between the N-terminus and the third zinc finger.
In the presence of inhibitory concentrations of Zn or Cd, ATP has been found to stimulate or re-activate the nuclease activity of CPSF30 (8). ATP stimulation requires the C-terminal 100 amino acids of the protein. This C-terminal domain apparently includes a hitherto unrecognized nucleotide-interacting domain, one that promotes nuclease activity. The significance of these properties - heavy metal inhibition and ATP stimulation of the nuclease activity - remains to be determined. One possibility is that they reflect regulatory interactions that link RNA processing with the heavy metal and metabolic status of the cell.
The two activities of the Arabidopsis CPSF30 are also affected in different ways by DTT. Specifically, RNA binding is relatively insensitive to the sulfhydryl reagent whereas nuclease activity is very sensistive (8). This sensitivity is due to the fact that two of the cysteine residues in the third CCCH motif are engaged in a DTT-sensitive disulfide linkage (9). This is an unusal finding, and suggests that the Arabidopsis CPSF30 is regulated in the cell by oxidation and reduction of this disulfide linkage. This possible link with redox regulation is intriguing, since the Arabidopsis CPSF30 has been linked with enhanced tolerance of plants to oxidative stresses (10). Along with the interaction with calmodulin, these properties indicate that CPSF30 is a link between different cellular signaling pathways and mRNA polyadenylation. Current research is focused on identifying the possible targets of this regulation.
Characteristics of an Arabidopsis mutant deficient in CPSF30 expression
Together with our collaborators (Drs. Li and Falcone), we are characterizing a mutant with a T-DNA insertion in the first exon of At1g30460, the gene that encodes CPSF30. While this mutant produces no detectable CPSF30 protein or mRNA (1), it is viable. Interestingly, this mutant is more tolerant than the wild-type to oxidative stresses (10). This mutant has several other phenotypes - it is deficient in lateral root production, it has a curious defect in fertility that is seen only in the first flowers produced after bolting, and it has altered responses to a number of plant hormones (11).
Using this mutant, we have explored the roles of the calmodulin-binding activity in the functioning of AtCPSF30. To do this, the smaller of the two proteins was expressed in the CPSF30 mutant. In addition, a variant of this protein in which the calmodulin-binding domain had been altered so as to be nonfunctional was also expressed in the mutant. The characterizations of these lines (11) revealed that the calmodulin-binding domain is required for the functioning of AtCPSF30 in lateral root development and the responses of the plant to auxins, cytokinins, and gibberellic acid, but not for normal fertility, oxidative stress susceptibiulity, or the responses of plants to ethylene. These results suggest that AtCPSF30 may differentiate between input signaling cues to yield varied physiological responses. The nature of additional possible signaling inputs is being explored. These ideas - that AtCPSF30 is a multifacted integrator of signaling inputs and phenotypic outputs - have been developed in recent review articles (12, 13).
We have used high throughput sequencing approaches to characterize poly(A) site choice in the AtCPSF30 mutant, as well as in lines that express either the wild-type or calmodulin-binding mutant forms of the samall protein depicted in Fig. 1. These studies reveal that numerous (>5000) individual poly(A) sites in Arabidopsis are affected by either the presence or absence of AtCPSF30 (14). Sites seen only in the mutant lacked the so-called Near-Upstream Element, suggestive of a novel class of poly(A) signal that is responsive to CPSF30. Interetsingly, both the wild-type and calmodumlin-binding CPSF30 mutant proteins restore wild-type poly(A) site profiles to the AtCPSF30 mutant (11). This indicates that the smaller of the two proteins shown in Fig. 1 is sufficient for normal poly(A) site choice in Arabidopsis, and that the calmodulin-binding domain itself is not involved in mRNA 3' end formation. (Of course, the effects of calmodulin on RNA binding noted above suggest that the first zinc finger motif is important for this process; this is currently being tested.)

Figure 1. Illustration of the Arabidopsis gene (At1g30460) that encodes CPSF30. The structure of the genomic DNA is shown at the top, and the two transcripts that are derived by alternative RNA processing below. Transcript "1" encodes CPSF30, while transcript "2" encodes a novel CPSF30-YT521B fusion protein.
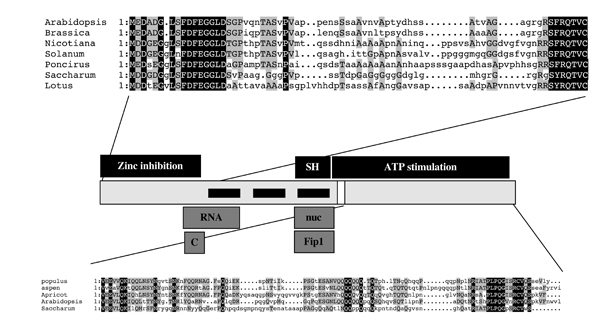
Figure 2. Functional map of the Arabidopsis CPSF30. The depiction in the middle illustrates the protein in linear fashion; thick black bars represent the three zinc finger motifs. Sequences above and below the depiction show alignments of the respective domains in plant CPSF30 proteins; positions of identity are denoted with while upper case lettering on a black background. Domains responsible for activities and interactions are noted above and beneath the depiction of the protein. SH - motif inhibited by DTT; RNA - RNA binding domain; C - calmodulin binding domain; nuc - zinc finger associated with nuclease activity; Fip1 - Fip1-binding domain.
1. Delaney KJ, Xu R, Zhang J, Yun K-Y, Li QQ, Falcone DF, Hunt AG. 2006. Calmodulin interacts with and regulates the RNA-binding activity of an Arabidopsis polyadenylation factor subunit. Plant Physiol. 140: 1507-1521.
2. Forbes KP, Addepalli B, Hunt AG. 2006. An Arabidopsis Fip1 homologue interacts with RNA and provides conceptual links with a number of other polyadenylation factor subunits. J. Biol. Chem. 281: 176-186.
3. Addepalli B, Hunt AG. 2007. A novel endonuclease activity associated with the Arabidopsis ortholog of the 30-kDa subunit of cleavage and polyadenylation specificity factor. Nucl. Acids Res 35, 4453-63.
4. Bai C, Tolias PP. 1996. Cleavage of RNA hairpins mediated by a developmentally regulated CCCH zinc finger protein. Mol Cell Biol 16: 6661-6667.
5. Bai C, Tolias PP. 1998. Drosophila clipper/CPSF 30K is a post-transcriptionally regulated nuclear protein that binds RNA containing GC clusters. Nucl Acids Res 26: 1597-1604.
6. Ryan K, Calvo O, Manley JL. 2004. Evidence that polyadenylation factor CPSF-73 is the mRNA 3' processing endonuclease. RNA 10: 563-573.
7. Mandel C, Kaneko S, Zhang , Gebauer D, Vethantham V, Manley JL, Tong L. 2006. Polyadenylation factor CPSF-73 is the pre-mRNA 3'-end-processing endonuclease. Nature 444: 953-956.
8. Addepalli B, Hunt AG. 2008. Redox and heavy metal effects on the biochemical activities of an Arabidopsis polyadenylation factor subunit. Arch Biochem Biophys 473, 88-95. doi:10.1016/j.abb.2008.02.027(request a reprint)
9. Addepalli B, Limbach PA, Hunt AG. 2010. A disulfide linkage in a CCCH zinc finger motif of an Arabidopsis CPSF30 ortholog. FEBS Lett. 584, 4408-4412. (request a reprint)
10. Zhang J, Addepalli B, Yun KY, Hunt AG, Xu R, Rao S, Li QQ, Falcone DF. 2008. A Polyadenylation Factor Subunit Implicated in Regulating Oxidative Signaling in Arabidopsis thaliana. PLoS One 3, e2410. (Open Access Article)
11. Liu M, Xu R, Merrill C, Hong L, Von Lanken C, Hunt AG, Li QQ. 2014. Integration of Developmental and Environmental Signals via a Polyadenylation Factor in Arabidopsis. PLoS ONE 9(12): e115779. (Open Access Article)
12. Hunt AG. 2014. The Arabidopsis polyadenylation factor subunit CPSF30 as conceptual link between mRNA polyadenylation and cellular signaling. Current Opinion in Plant Biology (Cell Signaling and Gene Regulation) 21C, 128-132. (request a reprint)
13. Chakrabarti M, Hunt AG. 2015. CPSF30 at the Interface of Alternative Polyadenylation and Cellular Signaling in Plants. Biomolecules 5(2), 1151-1168. (Open Access Article)
14. Thomas PE, Wu, X, Liu M, Gaffney, B, Ji G, Li QQ, Hunt AG. 2012. Genome-wide control of polyadenylation site choice by CPSF30 in Arabidopsis. Plant Cell 24, 4376-4388. (Open Access Article)
15. Hunt AG, Xing D, Li QQ. 2012. Plant polyadenylation factors: conservation and variety in the polyadenylation complex in plants. BMC Genomics. 2012 Nov 20;13:641. (Open Access Article)