MyoVision: automatic image quantification platform
Sections
1. About MyoVision
2. Core Features
3. Download
4. Documentation
1. About MyoVision
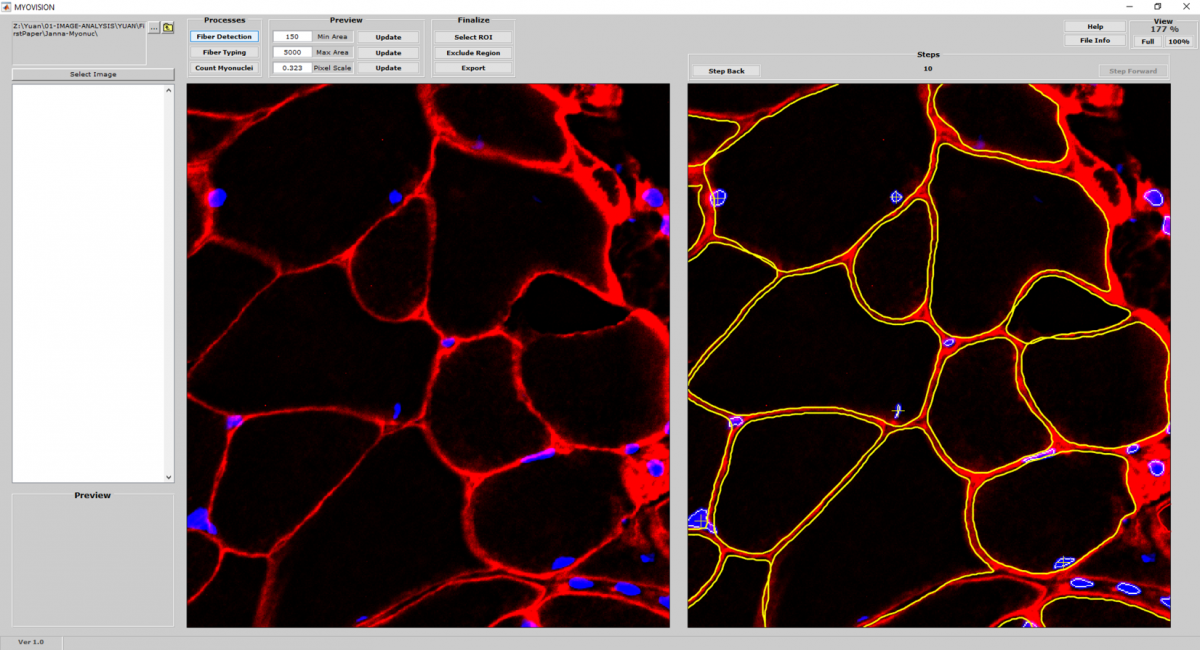
MyoVision is an automated image analysis progam to quantify muscle immunofluorescent microscopy images. Currently, MyoVision performs routine morphometry, including muscle fiber size, fiber type and myonuclear counting with minimal user input to reduce subjectivity and increase efficiency.
MyoVision incorporates and expands upon multiple algorithms specifically developed for skeletal muscle morphometry to achieve high degrees of accuracy and consistency*. MyoVision v1.0 is freely available for download (see section 3. below) and includes basic analytical features, supports limited user interactivity and is restricted to Microsoft Windows. The downloadable MyoVision v1.0 can handle only relatively small images, one at a time. MyoVision v2.0 is currently under development which will increase efficiency and accessibility, eliminating the requirement for users to have access to costly computer systems. This new release (later in 2019) will be a migration to open source, web-based software compatible with all operating systems, that will be hosted on servers at the University of Kentucky. MyoVision will serve as the foundation of a powerful analytical platform that will incorporate more advanced features, include quantification of additional muscle morphological characteristics, and expand multiplex analyses through implementation of user-friendly upgrades.
The Muscle Immunohistochemistry and Molecular Imaging Core (MIMIC) has high-performance parallel computing to enable MyoVision analyses of larger cross sections at higher magnification, and large numbers of images, with increased processing speed. Please contact us to discuss MIMIC performing high-throughput, automated quantification for your project. Images are analyzed in batches, data exported to an Excel spreadsheet provided to the investigator. Additionally, the investigator can view analyzed images for fiber outlines, fiber type classification and/or myonuclear counts using the MyoVision download. For questions or service requests please contact us.
*Wen Y, Murach KA, Vechetti IJ, Jr., Fry CS, Vickery CD, Peterson CA, McCarthy JJ, and Campbell KS. MyoVision: Software for Automated High-Content Analysis of Skeletal Muscle Immunohistochemistry. J Appl Physiol. jap 00762 02017, 2017.
2. MyoVision Core Features
Cell Detection and Counting
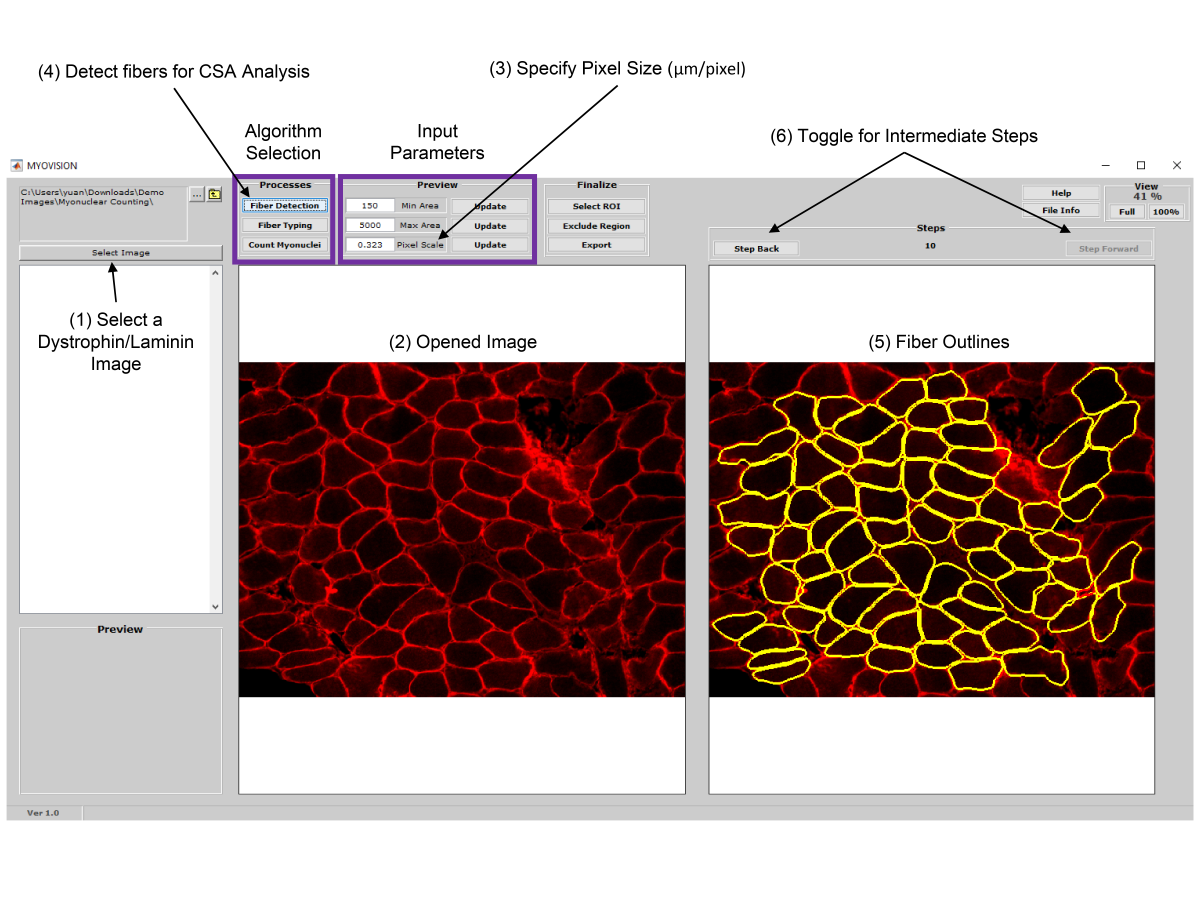
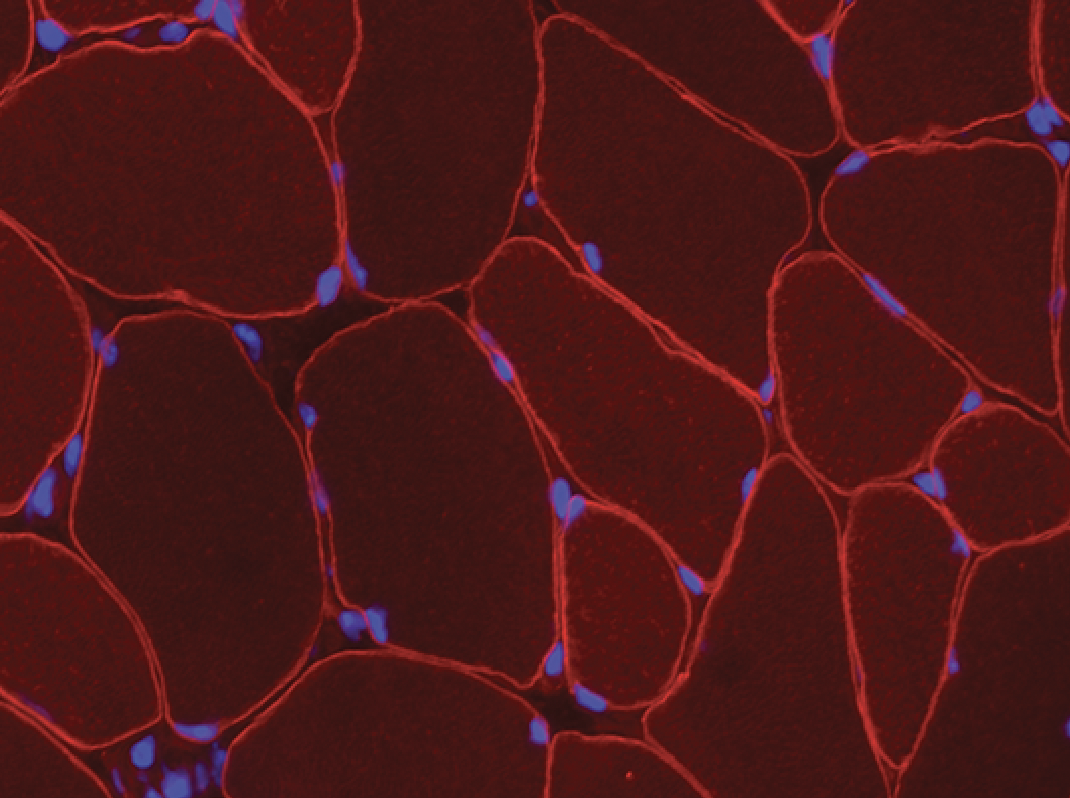
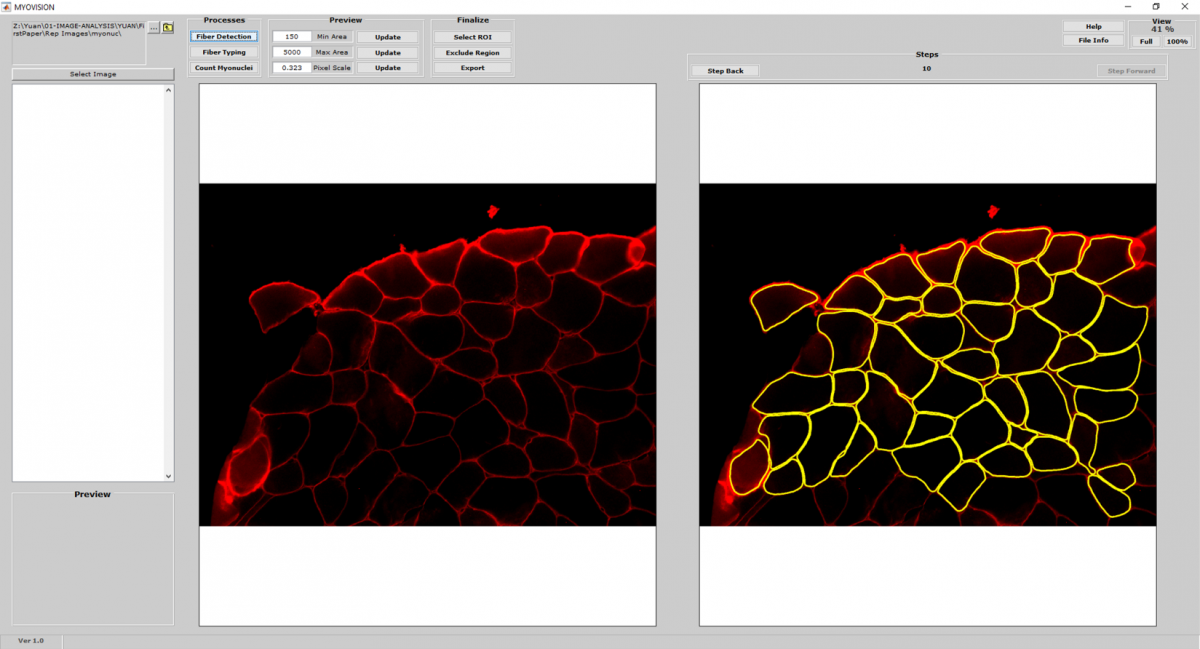
- MyoVision uses a laminin or dystrophin immunofluorescence of as a reference image to detect and count myofibers on cross section.
Cell Size Measurement
- Once myofiber cytoplasmic regions are detected and outlined, area and minimum Feret diameters are calculated for each cell.
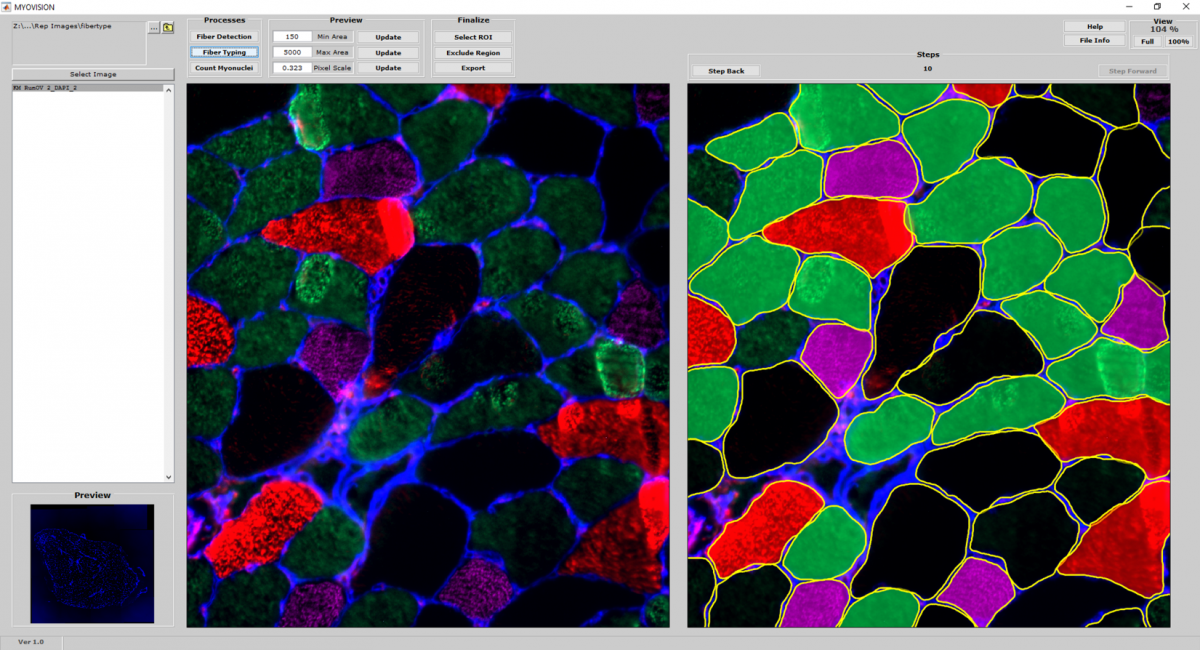
Fiber Type Classification
- Using the cytoplasmic outlines generated from the reference image as a mask, additional channels of myosin heavy chain immunofluorescence is quantified and used to classify fiber types.
Myonuclear Counting
- If the reference image is dystrophin immunofluorescence, the cytoplasmic mask can be applied to the DAPI channel to quantify myonuclear number.
3. Download
If you would like to download the MyoVision Installer Package, please fill out the webform at the following link: MyoVision Basic Downloads Form.
System Requirements:
Operating System: Windows 10
CPU: Intel i7-6700K
RAM: 16 GB (More if better)
Please note that MyoVision v1.0 is intended for the analysis of relatively small images one at a time in order to see the intermediate steps. Also, typical laptops and desktops may be too slow for large cross sections and high volume analyses. Please contact us to inquire about MIMIC analysis services using a dedicated high performance server (32 CPU cores with 250 GB of RAM).
4. Documentation
Quick-Start Guide
Fiber Type Demonstration Video