
Lecture Twenty Two
Nitrogen Metabolism: Source, Nomenclature, N2 Fixation

Goal: Introduction to the sources of nitrogen available to
plants, their chemistry and fixation into forms usable by plants. A good understanding of nitrogen fixation in plants.
Outline:
Background Readings for the discussion on N2 fixation:
a) REQUIRED:
1 - Chapter 16, sections 16.1 - 16.5 of the Biochemistry & Molecular Biology of Plants class text.
2 - Santoro, A.E. (2016). The do-it-all nitrifier. Science 351, 342.
b) SUGGESTED:
1 - Van de Velde et al. 2010. Plant Peptides Govern Terminal Differentiation of Bacteria in Symbiosis. Science 327:1122-1126.
2 - Hu, Y., A.W. Fay, C.C. Lee, J. Yoshizawa, and M.W. Ribbe. 2008. Assembly of Nitrogenase MoFe Protein. Biochemistry 47:3973-3981.
3 - Schrock, R.R. 2006. Nitrogen Fixation Special Feature: Reduction of dinitrogen. PNAS 103:17087.
4 - Tezcan, F.A., J.T. Kaiser, D. Mustafi, M.Y. Walton, J.B. Howard, and D.C. Rees. 2005. Nitrogenase Complexes: Multiple Docking Sites for a Nucleotide Switch Protein. Science 309: 1377-1380.
5 - Hipkin, C.R., D.J. Simpson, S.J. Wainwright, and M.A. Salem. 2004. Nitrification by plants that also fix nitrogen. Nature 430: 98-101.
Next to C, H and O, N is the most abundant element in plant tissues. In what form is most of the N on planet earth found?
The abundance (relative to 1000 atoms of C) of major elements
in plants and animals is illustrated in the following table:
| Element |
Zea mays |
Homo
sapiens (humans) |
| Hydrogen | 1705 | 2038 |
| Carbon | 1000 | 1000 |
| Oxygen | 765 | 252 |
| Nitrogen | 29 | 143 |
| Potassium | 6.5 | 6 |
| Calcium | 1.6 | 25 |
| Phosphorus | 1.8 | 22 |
| Magnesium | 2.0 | 1.4 |
| Sulfur | 1.5 | 5.2 |
| Chlorine | 1.1 | 2.8 |
| Iron | 0.4 | 0.05 |
As mentioned in the class text, Pliny the Elder ~ 80 A.D. stated "...It is however universally agreed that no manure is more beneficial than a crop of lupine turned in by the plough..."
Why is this?
Unlike O2 or CO2, plants (or any other eukaryote) cannot directly utilize N2 in biosynthetic reactions. The N2 must 1st be "fixed". Most fixed N in the biosphere is derived by biological N2 fixation by certain prokaryotes often in association with plants. Electrical energy from lightning can cause a small, but significant amount of N2 fixation, ~ 5 kg N ha-1 year-1. Until ~50 years ago agriculture depended on natural N2 fixation and recycling of previously fixed N.
| +3 | +1 | 0 | -1 | ||
| NO3-1 | NO2-1 | N2O2-2 | N2 | NH2OH | NH3 |
| nitrate | nitrite | hyponitrite | nitrogen gas | hydroxylamine | ammonia |
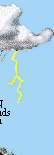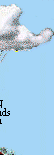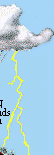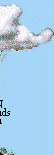
The N cycle is more generally illustrated in the following figure:
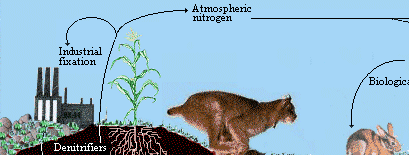 |
 |
 |
||
 |
 |
 |
Santoro (2016) described a do-it-all nitrifier.
Since then much of the N used in agricultural production is derived from industrial reduction of nitrogen to ammonia via the Haber-Bosch process started in 1913. In its simplest the equation for N2 fixation is:
 2NH3
2NH3The standard free energy, DG°' of this reaction is -33.5 kJ/mol indicating that this reaction is exergonic. This suggests that in the presence of a suitable catalyst this reaction would proceed spontaneously at room temperature. However, no catalyst is known for this to occur. Instead the Haber-Bosch process needs high temperatures (400-650°C) and pressures (100-400 atm) requiring considerable energy and expense. The synthetic nitrogen fixation consumes ~ 1/3 of the total energy for corn production (see Box 16.1 class text).
Biological N2 fixation:
N2 + 10H+ + 8e-  2NH4+ +
H2
2NH4+ +
H2
is also exergonic but in practice requires 16 molecules of ATP per molecule of N2 reduced. A source of e- is required and hydrogen gas is an additional product. 12-17 g of carbohydrate are consumed per g of N fixed.
The most efficient prokaryotic N2 fixation systems involve a symbiotic partnership with plants. There are at least 4 groups of such symbiotic interactions. Two involve soil bacteria and two involve cyanobacteria.
Various rhizobia genera and species are in the legume symbioses and Frankia bacteria in the actinorhizal symbioses. With the rhizobia and Frankia bacteria new structures are formed in the plant partners upon infection by the bacterial symbiont wherein the N2 fixation takes place.
The Nostoc cyanobacterium fix N in preexisting stem glands in Gunnera. Nitrogen can be supplied to rice paddies by the symbiosis of the water fern, Azolla, with the cyanobacterium, Anabaena.
In at least the first 3 cases the prokaryotes fix N as they reside inside their host cells. The symbiotic bacteria are released from infection threads into host cells in membrane-bound vacuoles termed symbiosomes. They are separated from their host cell cytoplasm by a peribacteroid membrane (PMB) derived from the plant host membranes.
Burkholderia...
The actual N fixation is carried out by the prokaryote and is catalyzed by an enzyme system known as the nitrogenase complex composed of dinitrogenase and dinitrogenase reductase. Nitrogenase and this process are highly sensitive to O2. The plant hosts provide 3 main functions in the symbiosis.

In what form does most of the N end up as in plant tissue?
The legume/Rhizobia symbiosis is the most important agriculturally. About 90% of the legumes examined to date have Rhizobium symbioses. In some legume crops N2 fixation can be quite substantial. The alfalfa/Rhizobia N2 fixation can be 150 to 200 kg N/ha per year and 100-150 kg N/ha with clover. Soybeans are the greatest N fixers among row crops and are reported to fix 50 to 150 kg N/ha. Common beans fix ~30 to 50 kg N/ha. Non-symbiotic N2 fixers fix less. In addition to fixing their own N, legume tissues tend to have higher N contents (protein) than other plants. Soybean seeds can be up to 50% protein.
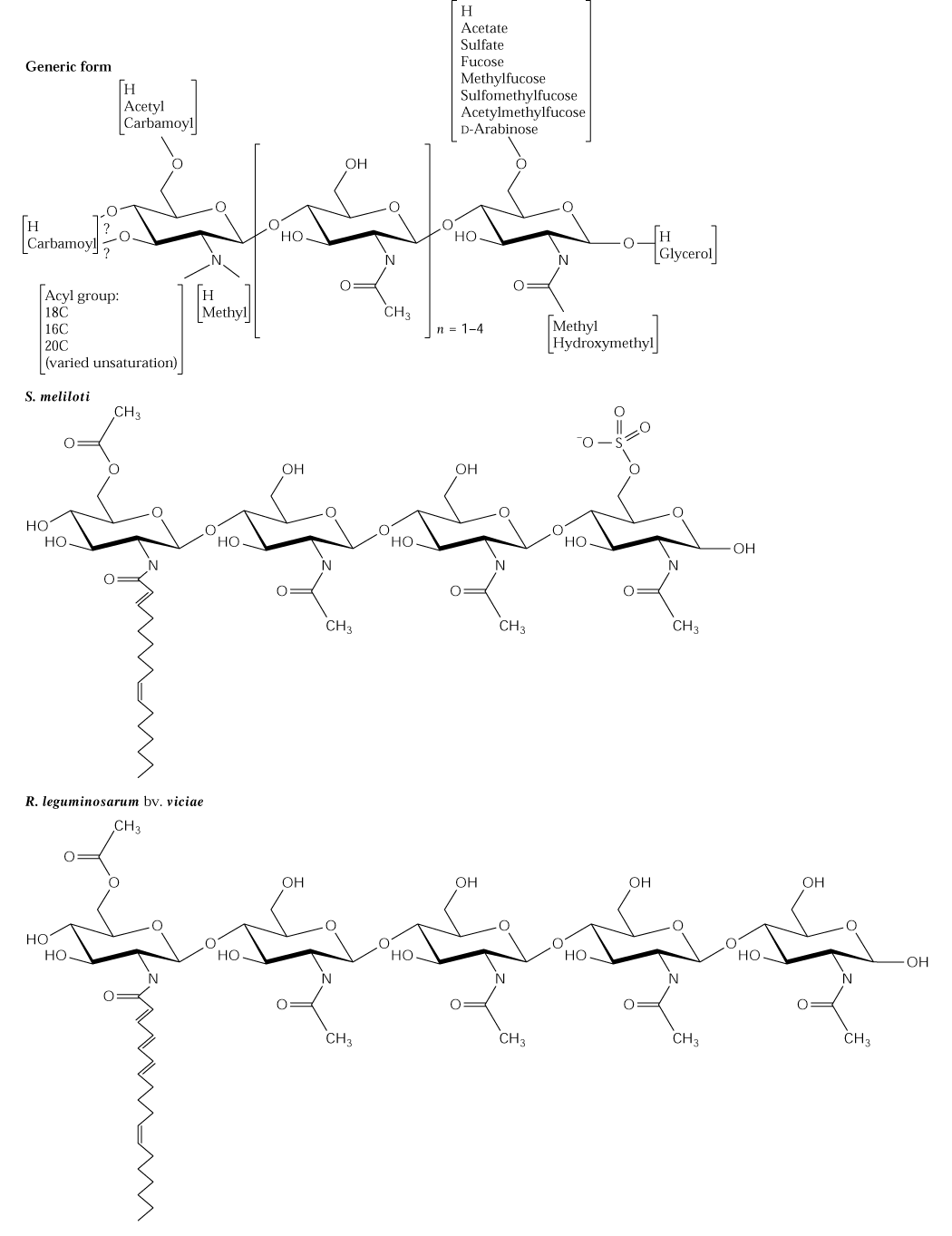
The formation of N2 fixing root nodules is known as nodulation and Rhizobia produce signal molecules known as Nod factors that play a key role in the initiation of nodulation. The host plant roots release specific flavonoid inducers of nod factors in the appropriate Rhizobial symbiont. The nod factors are lipochito-oligosaccharides with 3-5 ß-1,4-linked N-acetylglucosamine residues and a fatty acid (16:0-3, 18:0-1 or 18:4) on the non-reducing sugar monomer (Fig. 2; Mylona et al. 1995, The Plant Cell 7:869-885 or 16.24 of the class text).
Nitrogenase
Nitrogenase catalyzes the reaction:
N2 + 8H+ + 8e-  2NH3 + H2
2NH3 + H2
Each e- transfer requires 2 ATP molecules so the reaction includes:
16 ATP  16 ADP + 16 Pi
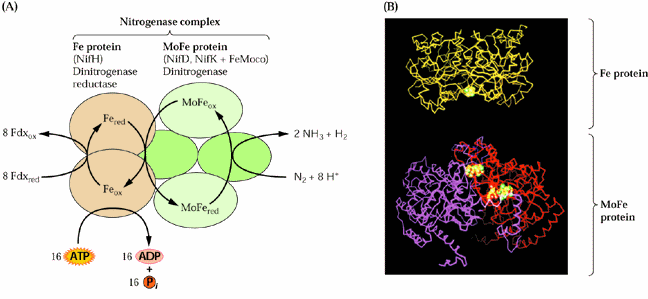
16 ADP + 16 Pi
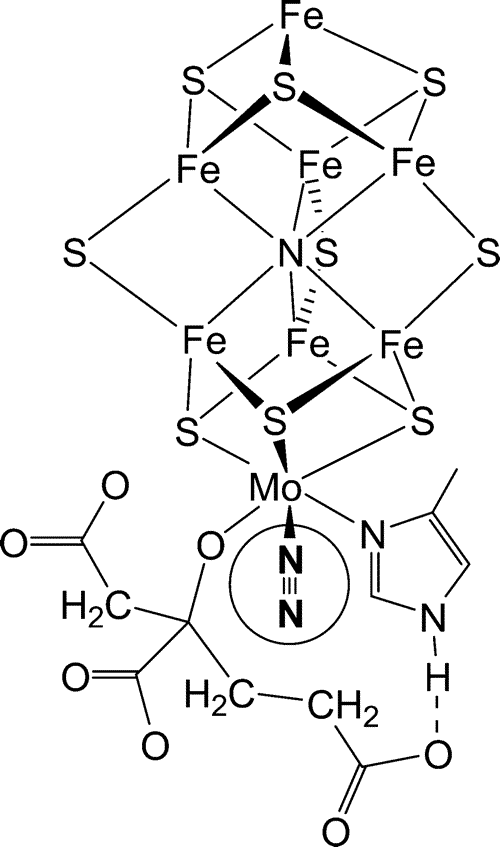
Nitrogenase enzymes contain an iron protein (dinitrogenase reductase) and a MoFe protein (dinitrogenase). The iron protein is composed of two ~65 kD O2 sensitive subunits containing 4 Fe and 4 S2- atoms per dimer. The MoFe protein is an a2b2 heterotetramer. The iron-molybdenum cofactor is a large redox center containing Fe4S3 and Fe3MoS3, which are linked to each other via 3 inorganic sulfide groups (see Fig. 11.7 of the Heldt text and 16.9 of the class text):
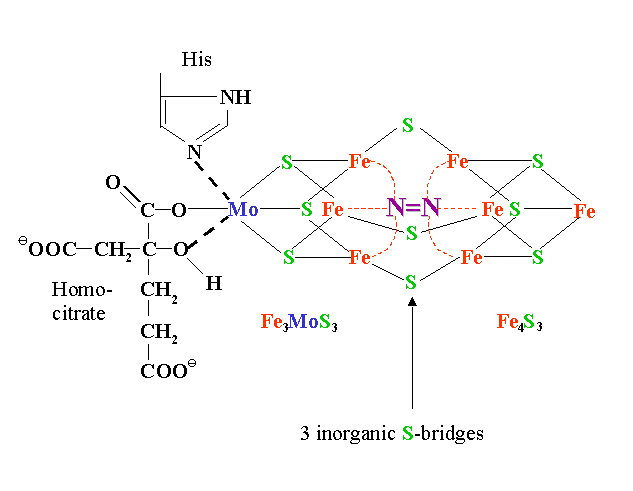
A model proposed by Seefeldt et al. (2004) for creation of a binding site for N2 at Mo by dissociation of the carboxylate ligand of (R)-homocitrate and hydrogen bonding of the longer arm of (R)-homocitrate to -442His (also see Weare et al., 2006).
The iron-sulfur center has a midpoint potential of -350 mV that increases to -450 mV when MgATP binds to the Fe protein and causes a conformational change. Upon reduction by NADH and binding MgATP, the Fe protein donates e- to the MoFe protein with concomitant hydrolysis of MgATP. It is the MoFe protein that carries out the N2 reduction. Thus the nitrogenase reaction can be divided into 3 steps:
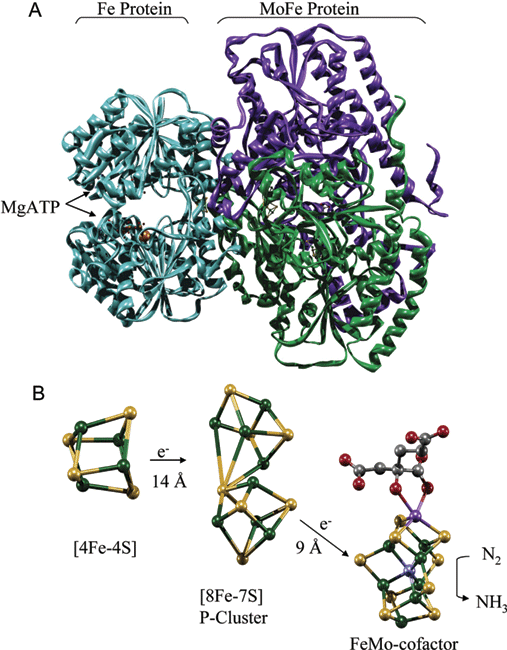
The overall nitrogenase reaction is:
This involves transfer of 8 e- from 4 NADH to N2 producing 2NH3 as illustrated in Fig. 11.6 of the Heldt text:
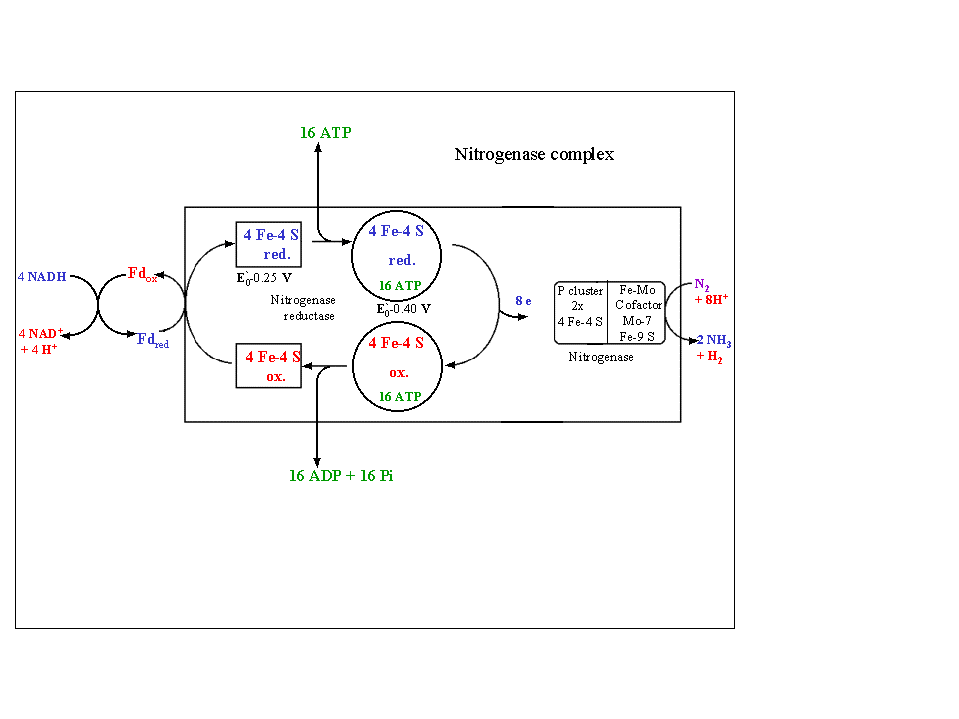
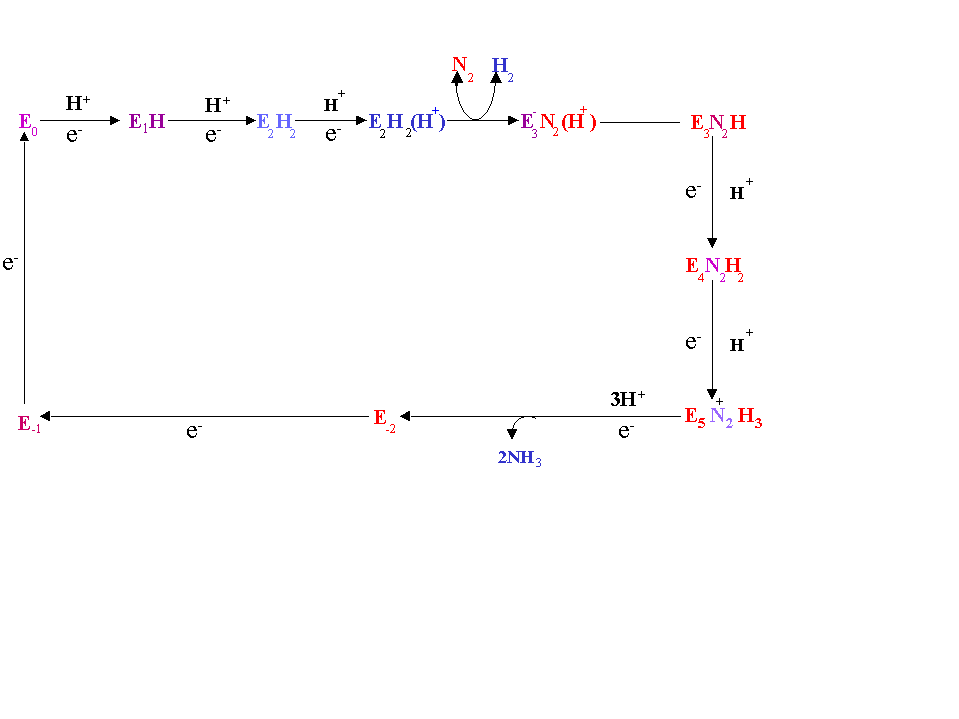
A model of this process is given in the following diagram (see also fig. 16.11 of the class text). The MoFe protein after being reduced by the Fe protein binds N2 (probably with participation of the Fe protein) releasing H2. Three e- are transferred to the N2 at the next step followed by 3 additional e- transfers upon which 2NH3s are released. The MoFe protein must be re-re-reduced by the Fe protein to accept another N2 molecule.
Nitrogenase can catalyze the reduction of other substrates with structures similar to dinitrogen. Acetylene reduction provides a convenient measure of nitrogenase activity:
HCCH + 2e- + 2H+  H2CCH2
H2CCH2
Another, generally more accurate (and difficult) way to measure N2 fixation is heavy N2, 15N2 incorporation:
15N2  15NH3
15NH3 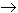 15N amino acids
15N amino acids
How is the 15N detected?
The H2 product of the nitrogenase reaction can be metabolized in some nodules. Some bacteroids possess hydrogenases which can reoxidize H2 to H2O:
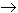 2H2O
2H2O
In other cases the H2 escapes into the atmosphere (and can actually be detected over clover fields).
Nodule function
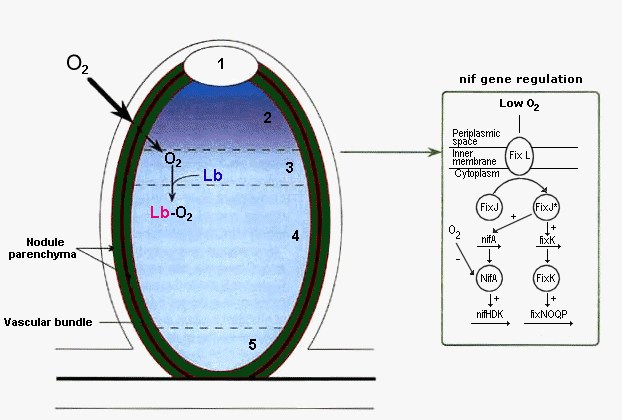
When bacteria are released from the infection threads into the infected cells, nodule primordia differentiate into N2 fixing nodules. Some nodules develop indeterminately in which a developmental gradient from the meristem to a senescence zone exists with the central tissue divided into specific zones (Fig. 4; Mylona et al. 1995, The Plant Cell 7:869-885). The nodule parenchyma (green) (red in Mylona et al. 1995). surrounding the nodule vascular bundle (black) provides an O2 barrier that reduces access of O2 to the central tissue. Since this is interrupted at the meristem, an O2 gradient is created dropping off toward the senescence zone. In the 1st interzone cell layer, the low O2 level activates the trans-membrane O2 sensor protein FixL. FixL activates the transcriptional factor, FixJ, by phosphorylation(*). FixJ* induces transcription of the nifA and fixK genes whose products in turn induce transcription of genes encoding the various proteins involved in the N2 fixation process. The NifA protein is also O2 sensitive providing an additional level of control of the process.
Van de Velde et al. (2010) report that plant peptides can govern terminal differentiation manipulating the cell fate of endosymbiosisbacteria. See Fig. 1. In some cases this symbiosis can result in "reverse parasitism"!
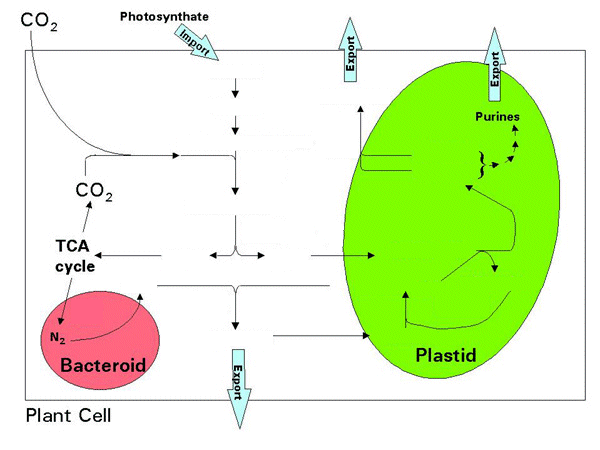
Sucrose transported from leaves provides the carbon source for nodules, which enters nodule metabolism after degradation by sucrose synthase. Carbohydrate metabolism supports the growth of the bacteriods while metabolism of dicarboxylic acids, e.g. malate, derived from the TCA cycle fuel N2 fixation.
O2 is transported to sites of respiration by leghemoglobin (Lb) encoded by the host plant. This can accumulate to 25% of the soluble protein of infected cells and confers a pink/red color characteristic of nodules active in N2 fixation. Like hemoglobin, leghemoglobin has a high affinity for O2 and controls delivery of O2 to respiratory enzymes while keeping O2 low in the vicinity of nitrogenase.
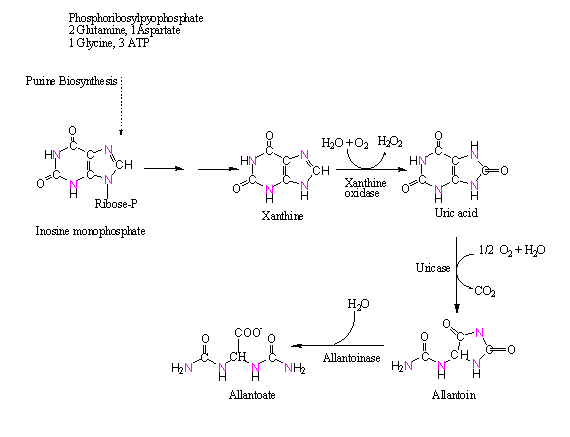
Legumes use 2 different pathways to transport fixed nitrogen out of nodules. The NH4+ produced in N2 fixation is 1st added to glutamate by glutamine synthetase forming glutamine (see below and next lecture). The amino group can then transferred from glutamine to oxaloacetate forming asparagine. In some legumes asparagine and glutamine are the principal form of transport of the fixed N throughout the plant via the xylem. In tropical legumes fixed nitrogen is often transferred from glutamine, via serine/glycine and aspartate, into purine biosynthesis in plastids of host cells. The purine xanthine is converted into uric acid in peroxisomes of surrounding uninfected cells of the nodules where it is converted into uric acid and then into allantoin and other N-rich ureides for transport via the xylem:
The ammonium produced via N2 fixation is assimilated into other nitrogenous
compounds as illustrated for a generalized legume in the following figure. Drag the
intermediate compounds and enzymes into the appropriate place in the figure from the
table at the right:
 |
|
The overall nitrogenase reaction is:
N2 + 4NADH+ H+ + 16MgATP  2NH3 + H2 +
4NAD+ + 16MgADP + 16Pi
2NH3 + H2 +
4NAD+ + 16MgADP + 16Pi
Nitrogen assimilation and photoautotrophic nitrification in legumes, Fig. 1 Hipkin et al. (2004).
Background Readings for the discussion on nitrate reduction and ammonia assimilation:
a) REQUIRED:
1 - Chapter 16, sections 16.6 - 16.10 of the Biochemistry & Molecular Biology of Plants class text.
2 - Han, Y.-L., Song, H.-X., Liao, Q., Yu, Y., Jian, S.-F., Lepo, J.E., Liu, Q., Rong, X.-M., Tian, C., Zeng, J., et al. (2016). Nitrogen use efficiency is mediated by vacuolar nitrate sequestration capacity in roots of Brassica napus. Plant Physiology.
b) SUGGESTED:
1 - Lee, J.-H., B.S. Evans, G. Li, N.L. Kelleher, and W.A. van der Donk. 2009. In Vitro Characterization of a Heterologously Expressed Nonribosomal Peptide Synthetase Involved in Phosphinothricin Tripeptide Biosynthesis. Biochemistry 48:5054-5056..
2 - Crawford, Nigel M. 1995. Nitrate: Nutrient and signal for plant growth. The Plant Cell 7: 859-868.
3 - Scheible, W.-R., R. Morcuende, T. Czechowski, C. Fritz, D. Osuna, N. Palacios-Rojas, D. Schindelasch, O. Thimm, M.K. Udvardi, and M. Stitt. 2004. Genome-Wide Reprogramming of Primary and Secondary Metabolism, Protein Synthesis, Cellular Growth Processes, and the Regulatory Infrastructure of Arabidopsis in Response to Nitrogen. Plant Physiol. 136: 2483-2499.
4 - Remans, T., P. Nacry, M. Pervent, T. Girin, P. Tillard, M. Lepetit, and A. Gojon. 2006. A Central Role for the Nitrate Transporter NRT2.1 in the Integrated Morphological and Physiological Responses of the Root System to Nitrogen Limitation in Arabidopsis. Plant Physiol. 140:909-921.
5 - Lea, U.S., M.-T. Leydecker, I. Quillere, C. Meyer, and C. Lillo. 2006. Posttranslational Regulation of Nitrate Reductase Strongly Affects the Levels of Free Amino Acids and Nitrate, whereas Transcriptional Regulation Has Only Minor Influence. Plant Physiol. 140: 1085-1094.
6 - Okamoto, M., A. Kumar, W. Li, Y. Wang, M.Y. Siddiqi, N.M. Crawford, and A.D.M. Glass. 2006. High-Affinity Nitrate Transport in Roots of Arabidopsis Depends on Expression of the NAR2-Like Gene AtNRT3.1. Plant Physiol. 140:1036-1046.
| New Version for 2017
All materials © 2017 David Hildebrand, unless otherwise noted. | |||||||
| home | syllabus | lecture schedule & web notes | virtual office hours | messages & answers from the instructor | supplementary material | related links | What's new? |